Bionty: Curate metadata using ontologies#
Look up records with auto-completion.
Map & curate metadata.
Manage public & custom ontologies and their versions.
To query, collaborate on, and persistently store knowledge & data, consider Bionty’s SQL interface with LaminDB - open-source data lake for biology.
Out-of-the-box ontologies#
Protein: Uniprot
Species: NCBI Taxonomy, Ensembl Species
CellLine: Cell Line Ontology
CellType: Cell Ontology
CellMarker: CellMarker
Tissue: Uberon
Disease: Mondo, Human Disease
Phenotype: Human Phenotype
Pathway: Pathway Ontology
Readout: Experimental Factor Ontology
Check out versions.yaml for details.
Installation#
Bionty is a Python package available for
pip install bionty
Import#
In your python script, import Bionty as:
import bionty as bt
Look up ontology terms with autocompletion#
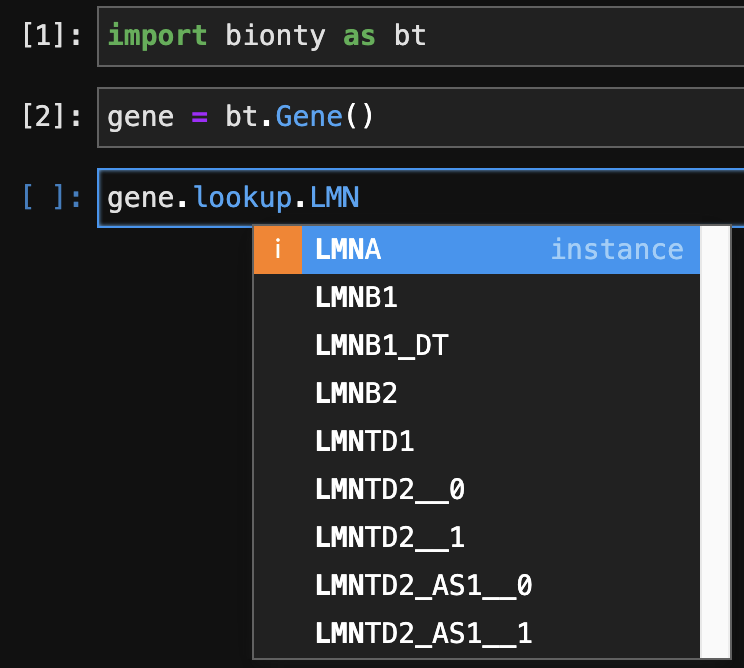
gene = bt.Gene()
gene.lookup.LNMA
See Look up records of species, gene, protein, cell marker for more.
Curate metadata#
import pandas as pd
# Create an example Pandas DataFrame of various cell types.
df = pd.DataFrame(
index=[
"placental epithelial cell",
"capillary",
"This cell type does not exist",
]
)
# The DataFrame can either be curated by ontology ID (id="ontology_id")
# or by ontology term names (id="name").
curated_df = bt.CellType(id="name").curate(df)
# ✅ 2 terms (66.7%) are mapped.
# 🔶 1 terms (33.3%) are not mapped.
See Curate entity identifiers for more.
Access ontology databases and versions#
# Display all managed versions
bt.display_available_versions()
# Access to the Mondo ontology
disease = bt.Disease(database="mondo")
# Access to the Human Disease ontology
disease = bt.Disease(database="doid", version="2023-01-30")
Didn’t see your favorite database or version? See how to Extend Bionty.


